Summary
Clinical Variant Ark (CVA) Portal is the visual gateway into the knowledge base built from rare disease cases and variants within the Genomics England Interpretation Platform.
You can access CVA Portal, using your existing Genomics England username and password here, cva.genomicsengland.nhs.uk
In this release we have focused on adding some pathogenicity "tags" to the variants in CVA so users can quickly identify whether their variant has been previously interpreted by another laboratory
New features / improvements:
- Changed the ordering of the HPO term list on the variant page so that the mostly frequent HPO terms are displayed first. Previously they were alphabetical (CZFE-503)
- Provided a warning message to users when a region search is performed that is greater than the current 1Mb maximum limit (CZFE-507)
- Added to the variant search results a summary of the ACMG classifications for that variant and highlight if there are any conflicts (CZFE-537)
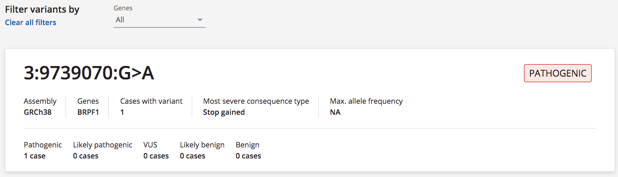
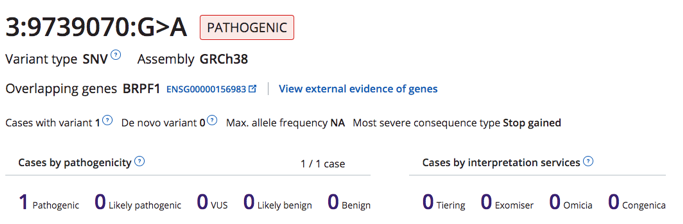
- Added the pathogenicity tag to the variant page (CZFE-536)
- Completed internal testing of the gms-beta instance of CVA so it is ready for NGIS BETA using AD credentials: https://cva-gms-beta.genomicsengland.nhs.uk
Fixed:
- Bug associated with clearing some of the case and variant filters (CZFE-591)
Known issues:
- It is not currently possible to filter variant search results by consequence type or pathogenicity, we are working on this for future releases
Access:
- CVA Portal BETA can be accessed on the HSCN network here: cva.genomicsengland.nhs.uk