Summary
Clinical Variant Ark (CVA) Portal is the visual gateway into the knowledge base built from rare disease cases and variants within the Genomics England Interpretation Platform.
You can access CVA Portal, using your existing Genomics England username and password here: cva.genomicsengland.nhs.uk
We have now made release notes and documentation for CVA available online here: https://cva-documentation.genomicsengland.co.uk/
New features and improvements
- Links to online release notes and user guide have been added to the footer accessible on every page
- You can now link directly to the Interpretation Portal from cases that are "To Be Reviewed"
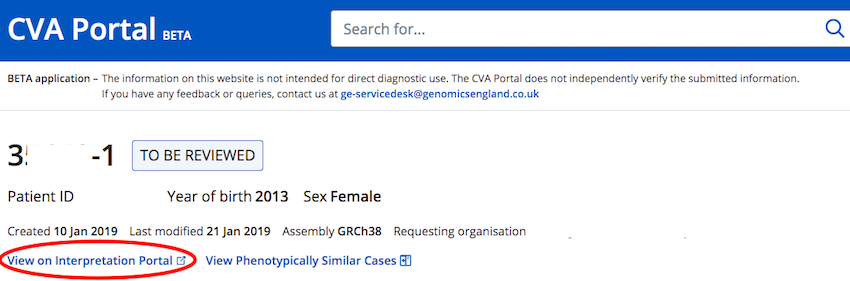
NOTE: You will only be able to review cases in the Interpretation Portal if you have permissions to do so i.e. if they are from your GMC
-
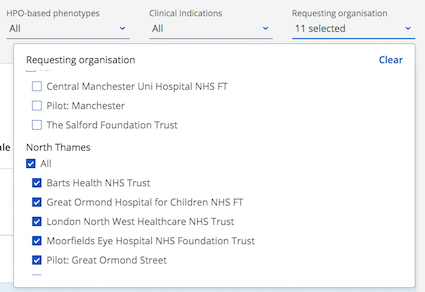
It's easier for users to filter by "requesting organisation" on the main search page and on the variant page.
The filter list has been updated to group organisations within a GMC
- The CVA Logo has been added to the site
Fixed
-
All Clinical Indications within a case are now being displayed
-
Variants that were previously missing from the "Summary fo Findings" and "Reported Outcomes Questionnaire" are now shown
-
Fixed missing gene and compound het filters not showing on variant table of case page
-
Case and Variant Filters are working for IE11
Known issues
- The de novo count on the variant page is counting report events not distinct variants so it is sometimes showing a de novo count higher than it should
- It is not currently possible to filter variant search results by pathogenicity; we are working on this for future releases
- Searching for genes will return cases where the case has a CNV called however, it is not currently possible to view CNV results in CVA
- Some variant pages are not rendered correctly because they are based on cancer germline variants which are in the CVA database but have a different data structure
Access
- CVA Portal BETA can be accessed on the HSCN network here cva.genomicsengland.nhs.uk