Summary
Clinical Variant Ark (CVA) Portal is the visual gateway into the knowledge base built from rare disease cases and variants within the Genomics England Interpretation Platform.
You can access CVA Portal, using your existing Genomics England username and password here: cva.genomicsengland.nhs.uk
New features and improvements
This release contains lots of updates to our Gene View page
You can access the Gene View page from any exisiting Case or Variant page in CVA by clicking on the HGNC symbol
PubMed articles have been added to the Gene View page
- Articles that have been included in the outcomes questionnaire for rare disease cases are now shown on the corresponding gene page of CVA

Table of reported variants has been added to the Gene View page
- The Gene View now displays a list of the variants that have an ACMG classification assigned from the reporting outcomes questionnaire
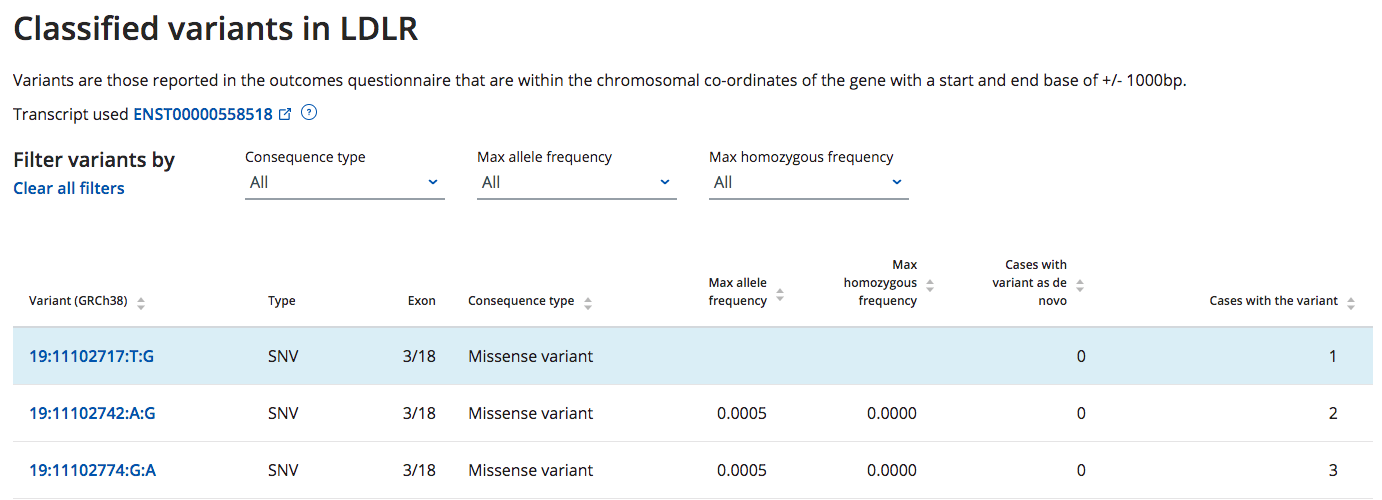
- Note the pathogenicity of each variant will be added in future updates
Case counts have been added to the Gene View panel table
-
The panel table on the Gene View now details the number of cases that have a ACMG classified variant in that gene when associated with a specific gene panel
e.g. using this table we hope users can identify potential new gene --> panel associations and update PanelApp evidence accordingly.
In the example below there is one case that has a reported VUS variant in the gene "ABHD12" which is currently "Low Evidence" on the Cataracts panel applied to the case
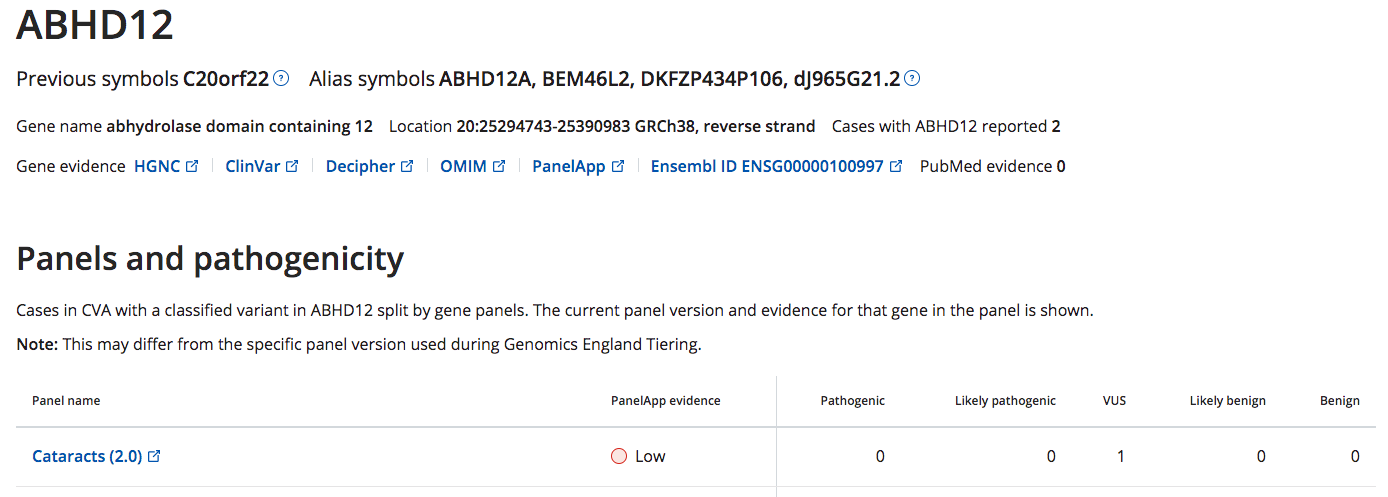
- Note the details of the cases in the table will be added in future releases via a side bar
A new filter has been added to the case list on all variant pages in CVA Portal
- The new filter is designed to reduce the chances of users identifying incidental findings in cases by first asking them to select a Clinical Indication they are interested in before being shown the list of cases with a particular variant
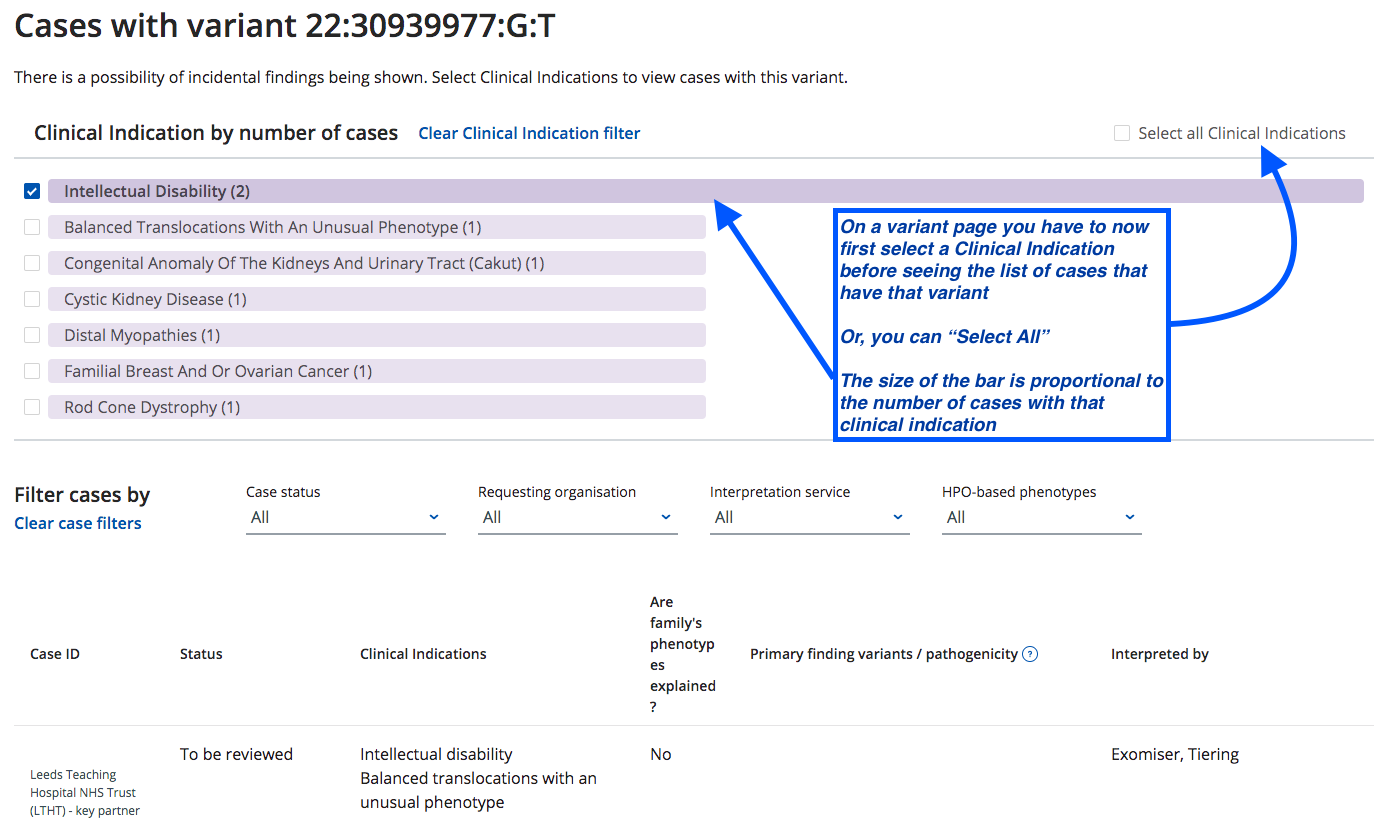
- Tabs now have labels depending on page you are on
Fixed
-
De novo variant count fixed so counting unique variants not report events.
Note currently the de novo count is only counting variants from Genomics England Tiering and not from other interpretation services such as Exomiser
-
The "Clinical Indications" filter on search results is now case insensitive
-
The overlapping gene tile on the variant view no longer displays "upstream" and "downstream" variants
-
Case level comments & variants from the summary of findings & reporting outcomes questionnaire are now displayed correctly in the sidebars
-
Variants from germline cancer cases which were rendering incorrectly in the Portal are no longer displayed
-
Link to the CVA API is fixed in the footer
-
Link to ClinVar improved to link to ClinVar search page rather than NCBI gene page
-
Minor updates to the text in the search bar and some of the tool tips
Known issues
- It is not currently possible to filter variant search results by pathogenicity; we are working on this for future releases
- Searching for genes will return cases where the case has a CNV called however, it is not currently possible to view CNV results in CVA
- Some variant pages are not rendered correctly because they are based on cancer germline variants which are in the CVA database but have a different data structure
Access
- CVA Portal BETA can be accessed on the HSCN network here cva.genomicsengland.nhs.uk