Summary
Clinical Variant Ark (CVA) Portal is the visual gateway into the knowledge base built from rare disease cases and variants within the Genomics England Interpretation Platform.
You can access CVA Portal, using your existing Genomics England username and password here: cva.genomicsengland.nhs.uk
New features and improvements
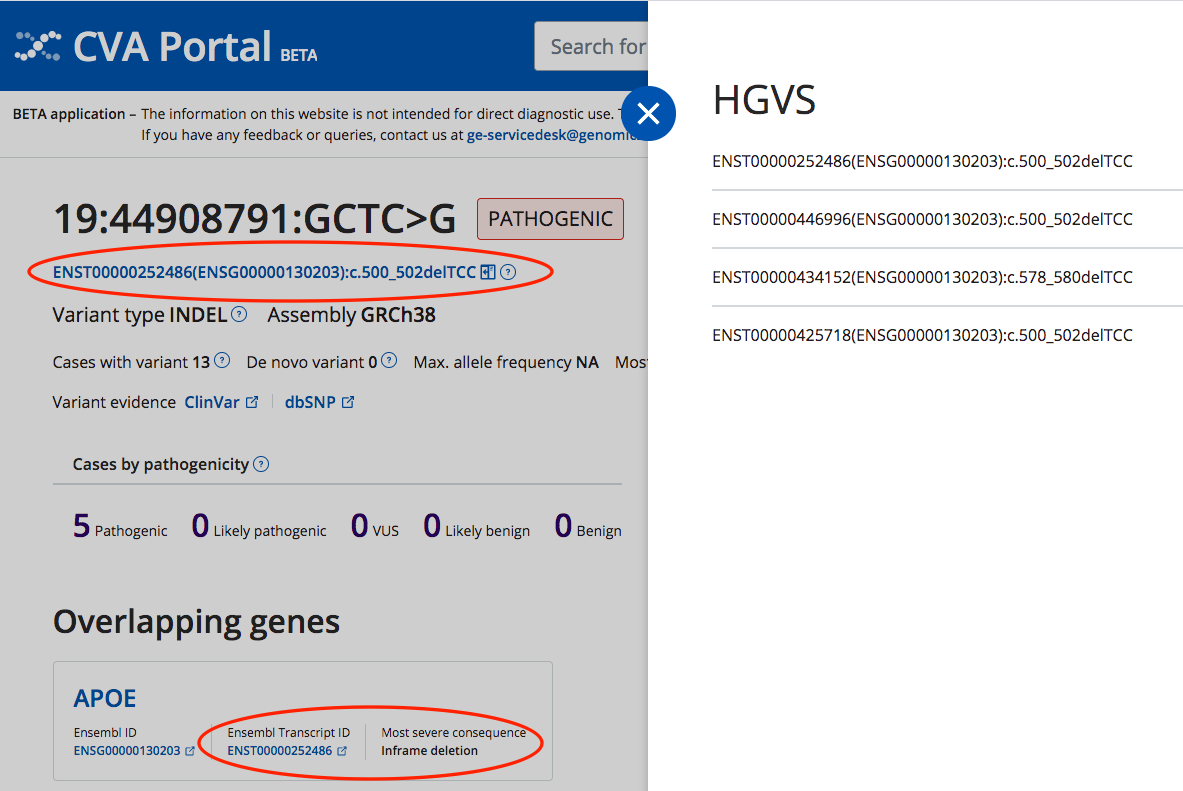
- c. HGVS variant annotation has been added to the CVA variant view page (CZFE-1118)
- Ensembl Transcript ID associated with the most severe consequence added to the gene card. (CZFE-1127)
- Improvements in load times of variant view pages (CZFE-1185)
Fixed
- Issue with the Interpretation Service variant count on the variant page is showing as zeros (CZFE-1173)
- Display of deletions impacting many nucleotides improved on the gene view page (CZFE-1142)
Known issues
- There is an issue with access to CVA Portal when your "session" expires. Currently, you do not get re-directed to the login page. This is causing the application to get stuck in a loop. The current workaround is re-open a fresh browser session. We are working to fix this ASAP (CZFE-1211)
- Variant, Case and Genomic Region Searches do not currently return a result in the Gene Search results tab
- It is not currently possible to filter variant search results by pathogenicity; we are working on this for future releases. The CVA gene page provides an overview of reported variants (those with a classification)
Access
- CVA Portal BETA can be accessed on the HSCN network here cva.genomicsengland.nhs.uk