Summary
Clinical Variant Ark (CVA) Portal is the visual gateway into the knowledge base built from rare disease cases and variants within the Genomics England Interpretation Platform.
You can access CVA Portal, using your existing Genomics England username and password here: cva.genomicsengland.nhs.uk
New features and improvements
- Support extended to Lab Sample IDs for FluidX tubes. This required migration of gelreport model to version 7.7.0 (CZFE-1472, CZFE-1480)
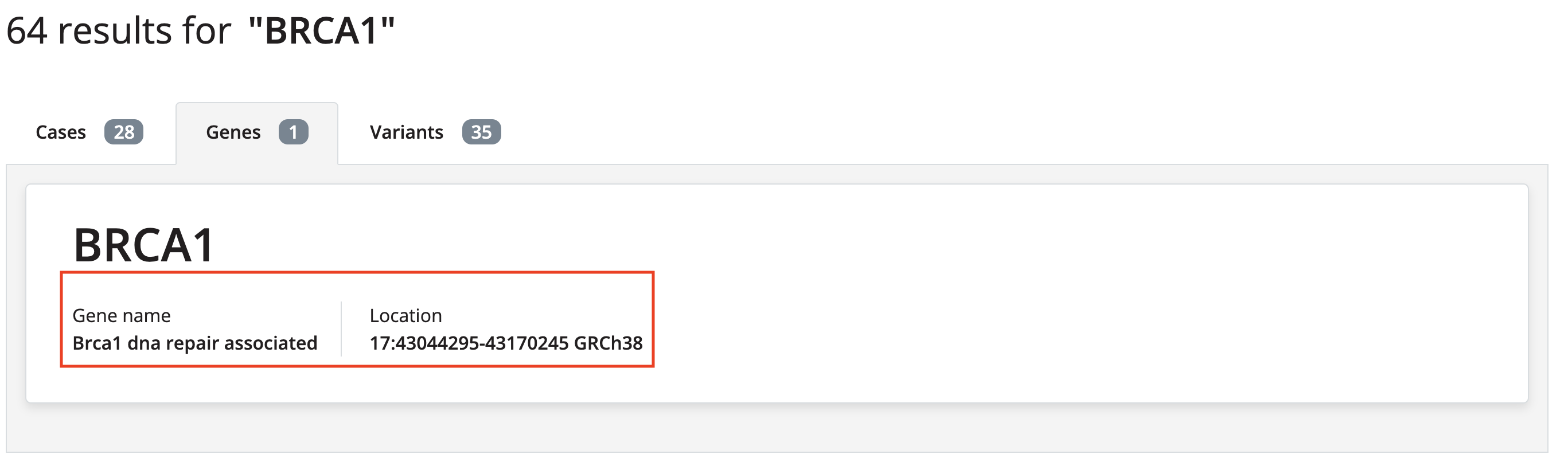
- Added additional gene information to the gene search card (CZFE-902)
-
The Variant Interpretation Logs (VILs) side bar on the variant list on the case page is now only displayed if there is a VIL available for that variant (CZFE-1305)
-
Added new VILs help section to online CVA documentation here
Fixed
- HPO term link out bug on case & variant pages (CZFE-1151)
Known issues
- Sending rare disease cases to Clinical Variant Ark (CVA) (CZFE-1555) We are investigating an edge case issue, where it appears that cases that are initially blocked in the CIP-API, then subsequently unblocked are not automatically being re-sent to CVA. This means, in some instances, users may observe rare disease cases not being available in CVA automatically. If you identify a GMS case missing from CVA please contact the service desk. We are working to improve this functionality in future releases.
- Variant, Case and Genomic Region Searches do not currently return a result in the Gene Search results tab
- It is not currently possible to filter variant search results by pathogenicity; we are working on this for future releases. The CVA gene page provides an overview of reported variants (those with a classification)
Access
- CVA Portal BETA can be accessed on the HSCN network here cva.genomicsengland.nhs.uk